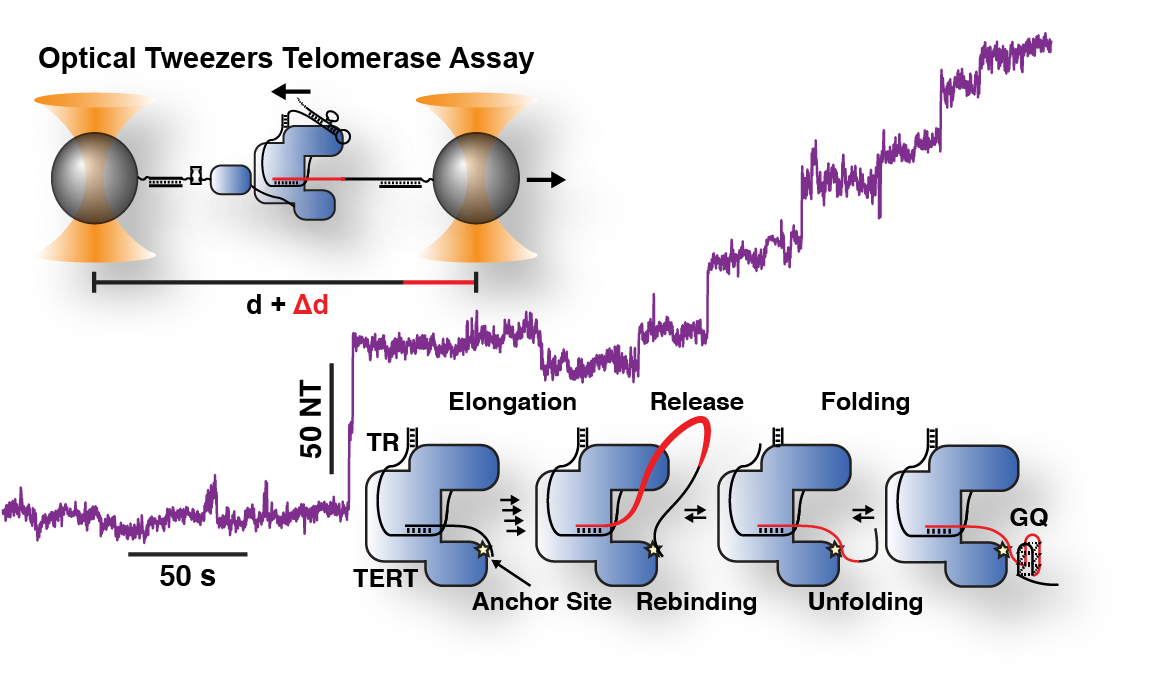
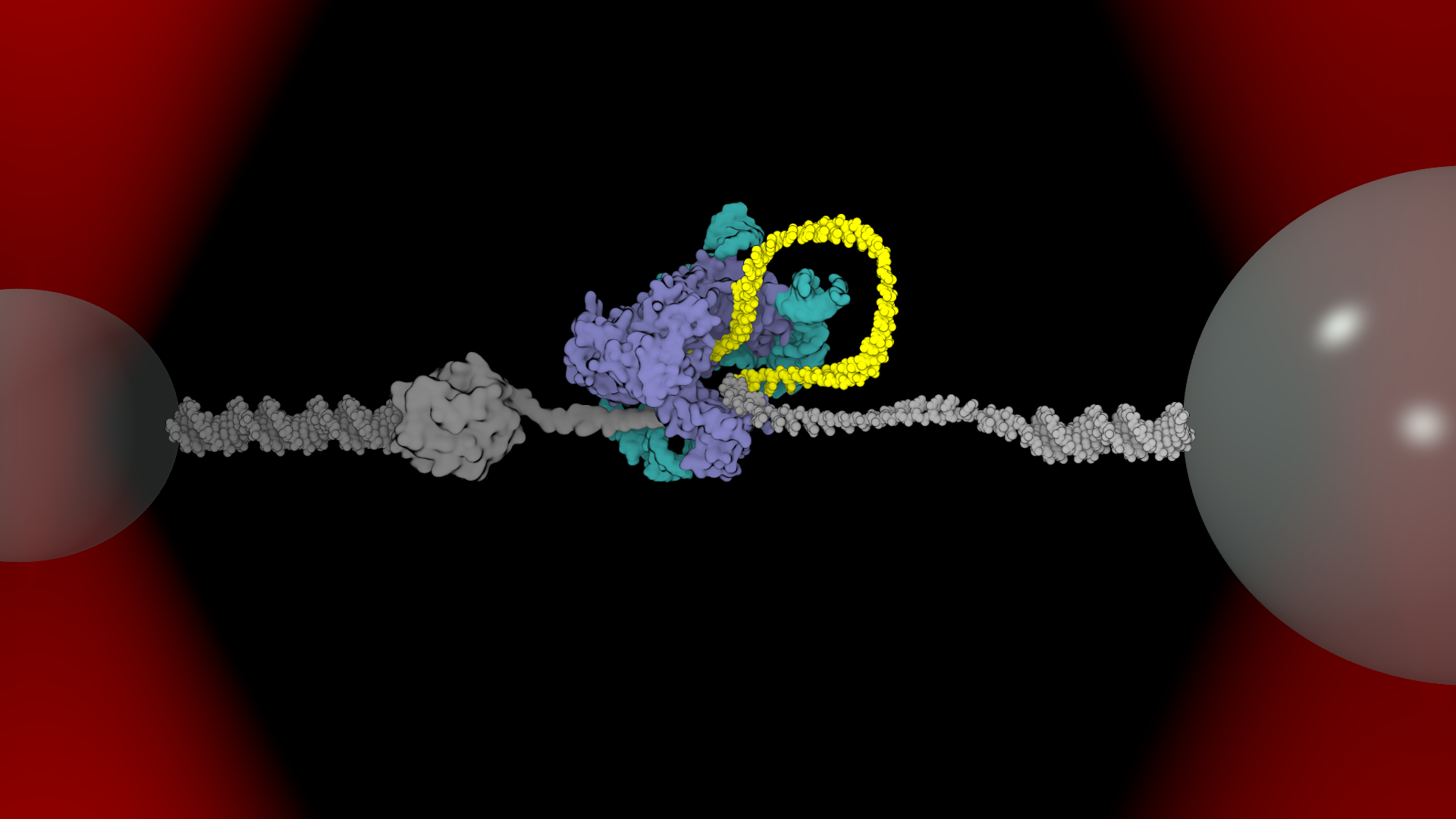
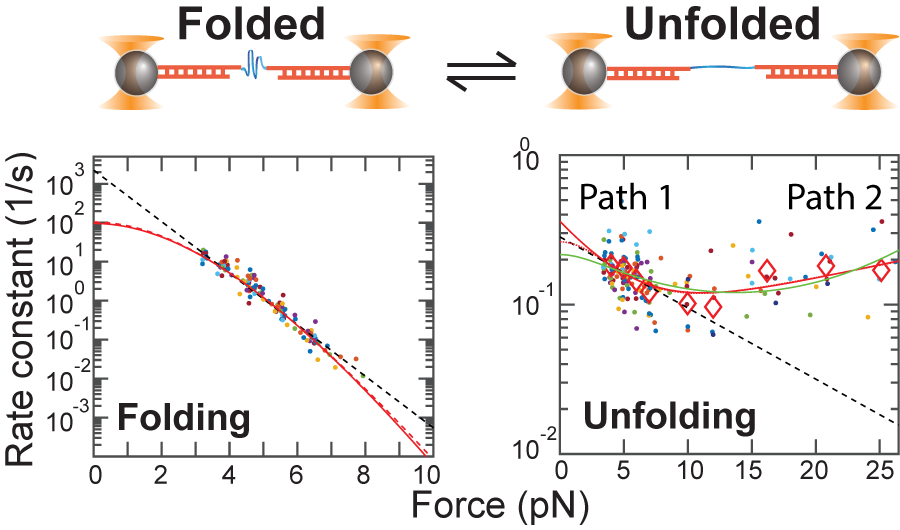
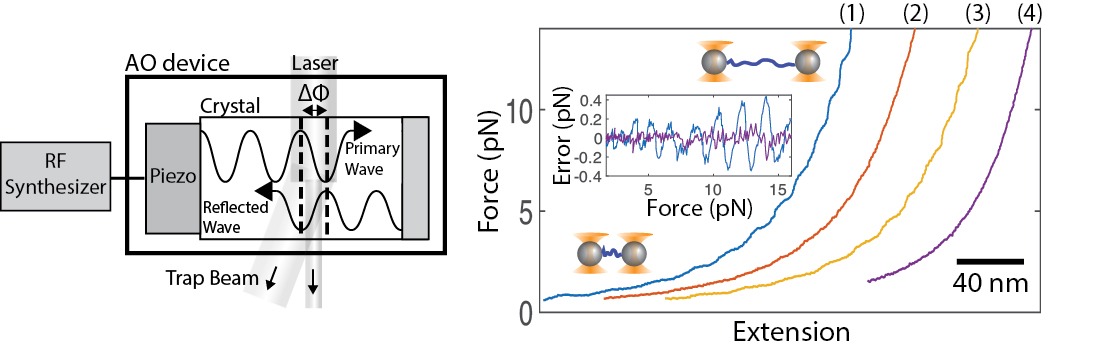
Lab Research Interest
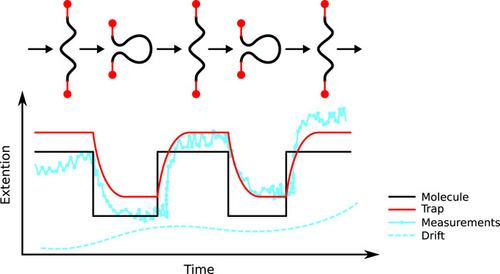
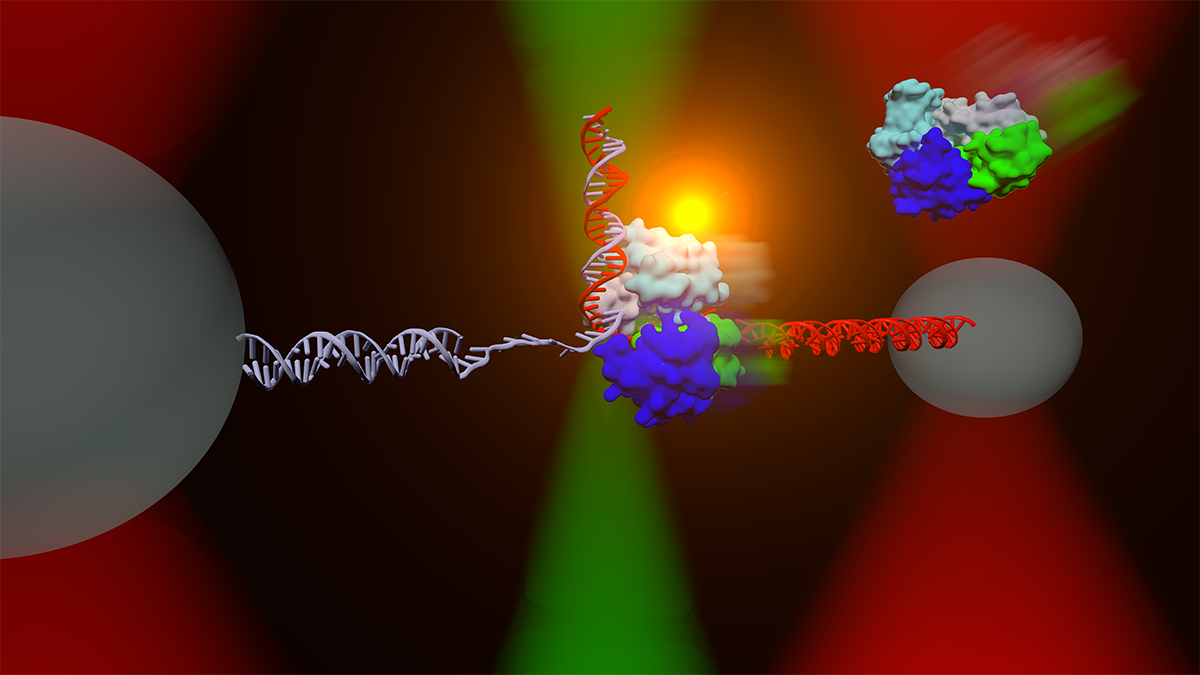
The Comstock lab investigates fundamental physical processes in biology using advanced, precision single-molecule measurement and manipulation techniques. We observe in real-time individual protein molecular machines marching down DNA strands or ripping open RNA. We manipulate live cells and measure individual cellular electrochemistry. To do this we design and build frontier biophysical instrumentation combining optical tweezers and single-molecule fluorescence microscopy. We strive to watch biology in action without the obscuring effects of traditional ensemble methods.
We are a young interdisciplinary lab composed of scientists with a diversity of backgrounds. Students have the opportunity to participate in all aspects of biophysical research including: wet lab development and production of biological systems, design and construction of instrumentation (optics, electronics, software etc.), quantitative data analysis and modeling and collegial discussion. We are always on the lookout for excited new colleagues. Interested students and postdocs with a background in physics, biology, chemistry, or a related field are welcome to contact Prof. Comstock.
News
April, 2022 MSU SciFest is back with in person on campus events including our very popular lab open house event "Optical Tweezers: Reach Out and Grab a Bacteria"! Our lab opened to the public and guests got to run our laser tweezers microscopes and hunt down two contrasting kinds of swimming bacteria and take a selfie with their (temporary) pet bacteria (thanks to Michaela TerAvest and Yann Dufour's labs for collaborating).
July, 2020 Due to the hard work and creativity of many previous and current members of the lab, Prof Matt Comstock has been promoted to Associate Professor with tenure. Thanks so much to the lab, MSU, the Jerry Cowen endowed chair, the NSF and members of Matt's family especially. A masked and distanced ceremony was held in 2022. Photo: Matt with MSU President Samuel Stanley Jr. and Provost Teresa Woodruff.
January 3, 2019 Congratulations to new PhD, Dr. Cho-Ying Chuang, our trapping lab's second successful PhD thesis!
Cho-Ying was a major asset to the lab, helping to build the lab and develop new fluorescence and tweezers methods. Among other projects, she made very high resolution measurements of very small nucleic acids folding (stay tuned!).
October, 2018 Together with the Lapidus lab (here at MSU), we have published a high-resolution force spectroscopy investigation of the folding and unfolding of a model protein, GB1. We performed equilibrium measurements down to very low forces and observed a clear transition from a high force mechanical unfolding mechanism to a low force more 'native-like' unfolding mechanism - a transition that has been seen only rarely previously. The high force results agree with previous out-of-equilibrium high pulling force results while the low force results seem to extrapolate much closer to ensemble results. This work highlights the potential for the coexistence of multiple folding and unfolding pathways and how they can mask each other in different force ranges. This reinforces the need to take care when extrapolating force spectroscopy data to zero force. We published our work in the recent 'Festschrift' honoring Bill Eaton:
"Combined force ramp and equilibrium high-resolution investigations reveal multi-path heterogeneous unfolding of protein G," Journal of Physical Chemistry B, 122, 11155 (2018).
February, 2018 We have published a method to completely vanquish optical trap positioning and measurement errors, known as 'wiggles', arising from acousto optic devices! These errors have long plauged high resolution methods in particular. While some workarounds exist, we show how simply reducing the coherence of the RF drive signal can completely remove the wiggles. This enhances tweezers stability and accuracy particularly for long measurement durations. The new 'random phase' method can be implemented via a software change in the RF synthesis method - no new equipment is necessary. Our latest high-resolution fleezers instrument software which incorporates this upgrade is now available for download.
"Randomizing phase to remove acousto-optic device wiggle errors for high-resolution optical tweezers ," Applied Optics 57, 1752 (2018).
October 26, 2017 Congratulation to newly minted Dr. Dena Izadi, our trapping lab's first successful PhD thesis!
Dena investigated protein folding complexity at high resolution and was co-advised by Prof. Lisa Lapidus.
January 25, 2017 Our lab's single molecule optical trapping and fluorescence methods are described in two recently released book chapters:
"High-resolution optical tweezers combined with single-molecule confocal microscopy," in “Methods in Enzymology”: “Single-Molecule Enzymology: Nanomechanical Manipulation and Hybrid Methods”, Spies, M. & Chemla, Y. R., (editors), Elsevier. 582 (2017).
"
"High-Resolution “Fleezers”: Dual-trap Optical Tweezers Combined with Single-Molecule Fluorescence Detection," in: “Optical Tweezers: Methods and Protocols”, Generich, A. (editor), Springer, (2016, in press).
These protocols and methods are provided in collaboration with Yann Chemla's lab at UIUC.
September 1, 2015 Our technique has been featured again in Genetic Engineering and Biotechnology News (GEN): More Dynamic Protein Profiling. See on page 2: Single Molecule Nanometry.
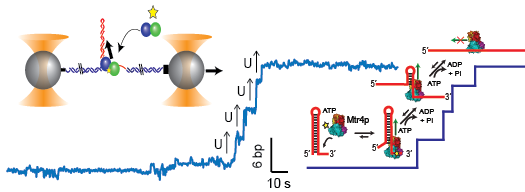
June 28, 2015 We have received our first extramual funding! We have received a regular NSF award for "Investigation of RNA-Processing Protein Dynamics with Simultaneous High-Resolution Optical Traps and Single-Molecule Fluorescence" (MCB-1514706).
April 17, 2015 Matt's postdoctoral work (Chemla and Ha labs) is published in Science!
M.J. Comstock, K.D. Whitley, H. Jia, J. Sokoloski, T.M. Lohman, Taekjip Ha, and Y.R. Chemla. “Direct observation of structure-function relationship in a nucleic acid-processing enzyme,” Science, 348, 352 (2015).
Here we directly observe the dependence of helicase unwinding activity on protein stoichiometry and conformation.
Support
We are presently supported by the National Science Foundation (MCB-1919439), MSU startup funds and the Cowen Endowed Chair which was made possible by the generous support of Randy Cowen and his family.