Molecular machines
Nanometer-scale protein molecular machines are at the core of all living systems. For example, helicase motor proteins consume ATP fuel molecules and to walk down DNA tightropes unwinding the double helix. This activity is essential for e.g., allowing access to the information stored in DNA or repairing damaged DNA. How these machines function remains mysterious.
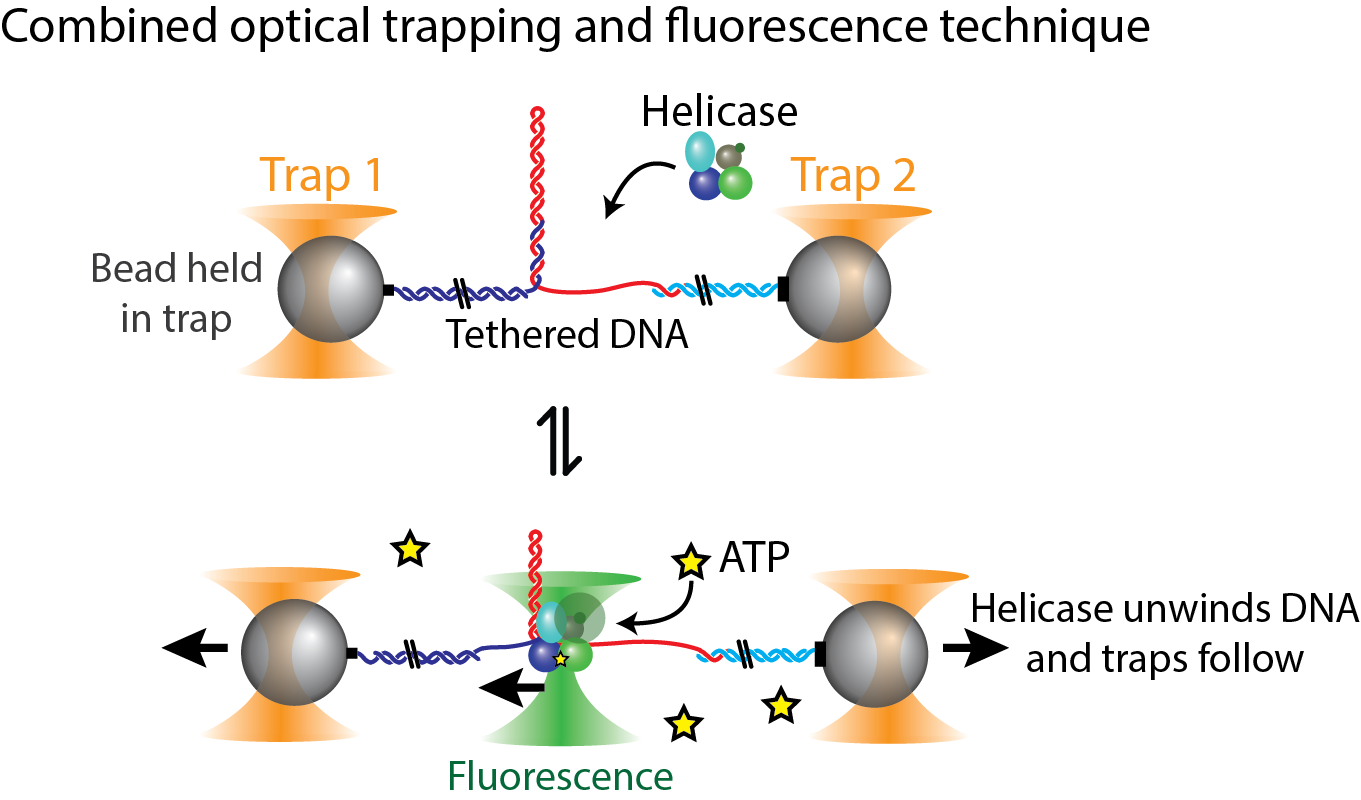
The Comstock lab investigates such processes using advanced, precision single-molecule measurement and manipulation techniques. Using optical traps we can grab and stretch a single DNA strand and observe the activity of individual protein molecular machines in real-time (see cartoon on right).
Amongst other projects, we are presently collaborating with the Jankowsky lab at Case Western Reserve University to investigate molecular machine processing of RNA.
We have recently published results from this project where we have directly shown that the nuclear RNA helicase Mtr4p (both alone and within the TRAMP RNA surveilance complex) is a slow translocase that stalls without an upstream duplex to unwind:
More recently we have been collaborating with the Schmidt lab here at MSU to investigate the human telomerase RNA + protein molecular machine. We have successfully developed a new high-resolution tweezers method that allows us to directly observe human telomerase processively (continuously) elongating the ends of DNA. We have demonstrated the existence of a telomerase anchor site that allows it to retain its hold on the DNA while its RNA template disengages and reorients. We have a preprint online of our manuscript presently under review:
For a basic overview of our technique see:
For detailed protocols, methods and new innovations:
Our multicolor high-resolution fleezers instrument: C.-Y. Chuang, M. Zammit, M. L. Whitmore, and M. J. Comstock, "Combined high-resolution optical tweezers and multicolor single-molecule fluorescence with automated single molecule assembly line," Journal of Physical Chemistry A, X, X (2019), in press.
Our 'random phase' method to drive the tweezers AOM to obliterate AOM 'wiggle' errors: A. G. Baker, C.-Y. Chuang, M. L. Whitmore, and M. J. Comstock, "Randomizing phase to remove acousto-optic device wiggle errors for high-resolution optical tweezers ," Applied Optics 57, 1752 (2018).
In collaboraion with Steve Pressé's group, a machine learning method to 'discover' states in a high-resolution tweezers time trajectory in an unbiased way: I. Sgouralis, M. L. Whitmore, L. J. Lapidus, M. J. Comstock, and S. Pressé, "Single molecule force spectroscopy at high data acquisition: A Bayesian nonparametric analysis," Journal of Chemical Physics 148, 123320 (2018).
Protein folding
The optical traps can be configured to tug on the ends of individual proteins to induce the unraveling and refolding of their structure. We are presently collaborating with the Lapidus lab to investigate protein folding processes using our novel high-resolution trapping and fluorescence capabilities.
The high-resolution of our tweezers allows us to directly measure protein folding and unfolding at very low forces, under 'gentle' equilibrium conditions. We recently showed that under such conditions, folding pathways can shift from mechanical (pulling) dominated to more native-like thermal pathways.